|
PAUL THOMPSON'S
CURRENT MATHEMATICAL/ENGINEERING PROJECTS
(for NEUROSCIENCE projects, click here) |
|---|
| TENSOR BASED MORPHOMETRY |
|---|
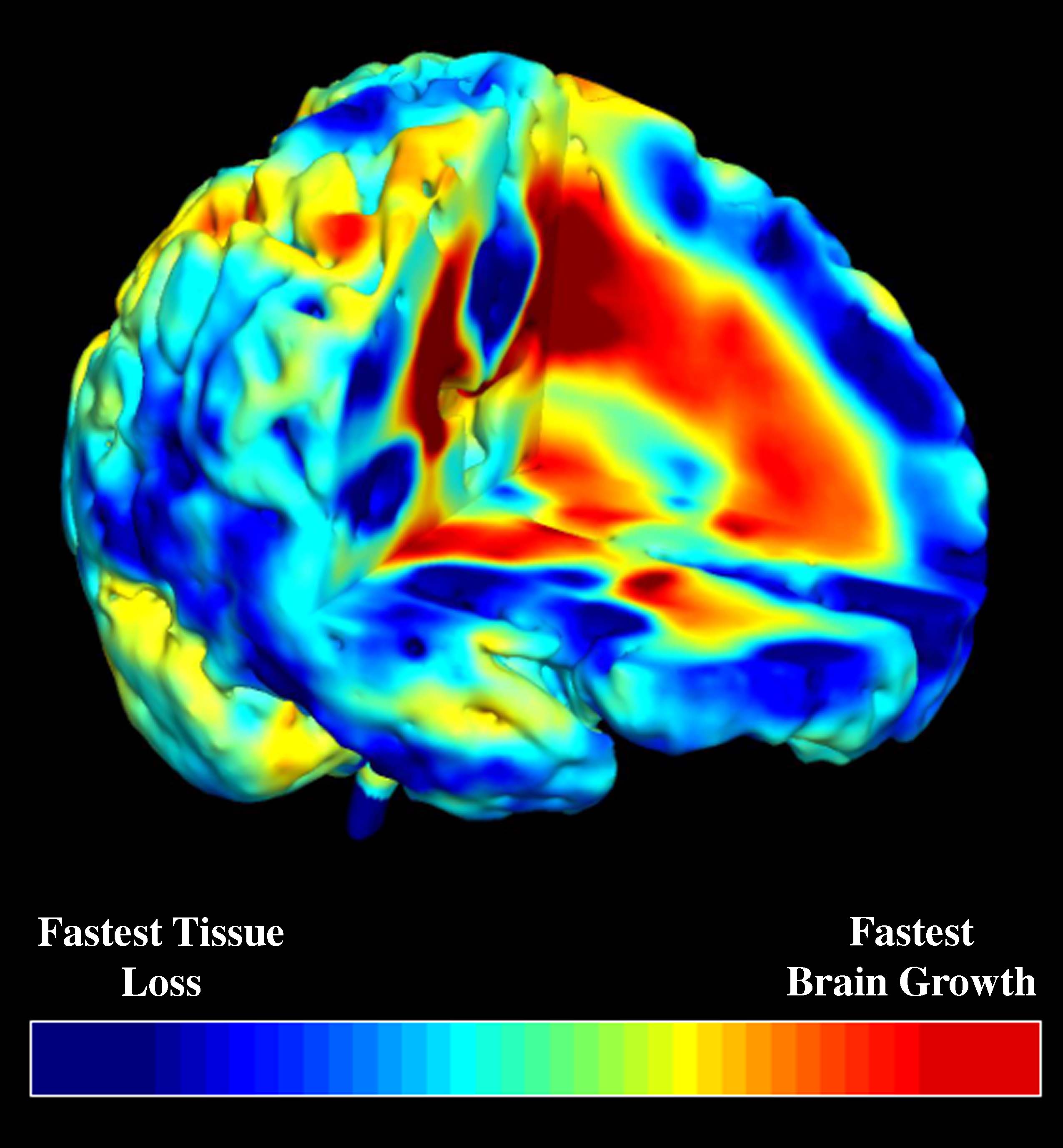




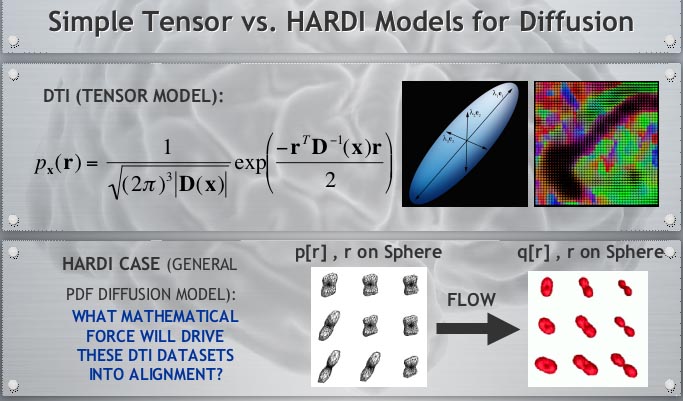
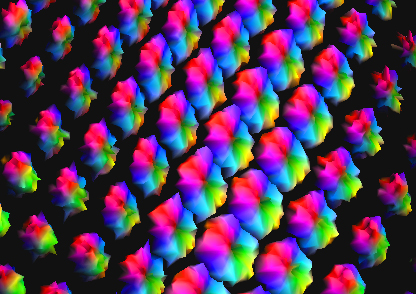
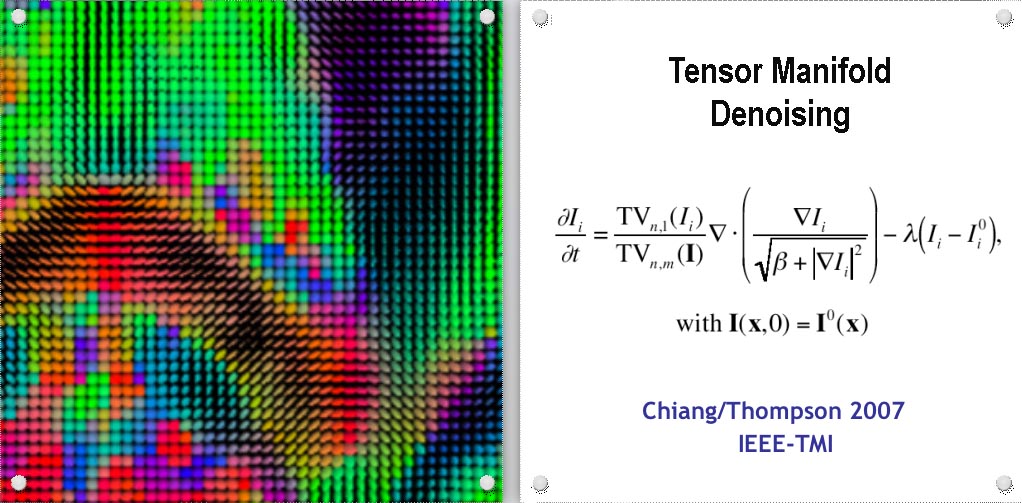
To track brain changes, during brain development and disease, and in drug trials, we are developing powerful tensor-based morphometry approaches to detect brain structure differences, using mathematics adapted from continuum mechanics, i.e. the analysis of elastic and fluid deformations. We also develop non-linear image registration approaches, which fluidly deform 3D brain images to match each other. The structural differences between HIV/AIDS patients' brains and healthy normal subjects' brains are detected here (red colors; 3rd image) by computing flow fields that deform one brain scan onto another, measuring brain anatomical differences in 3D. The same approach can map brain growth (1st image) or Alzheimer's Disease (2nd image). The applied deformation vector fields, with billions of degrees of freedom, are designed to align images in exquisite detail by maximizing the mutual information between their intensity distributions. We are exploring other information-theoretic approaches for image registration based on generalized divergences between probability distributions, such as the Jensen-Renyi divergence and Kullback-Leibler divergence on material density functions. These deformation fields can be analyzed statistically with Lie group methods, and log-Euclidean metrics, that operate on the manifold of symmetric matrices. Related work is developing nonlinear registration methods for alignment, denoising and statistical analysis of diffusion tensor images (see below). We are also studying the statistics of deformation tensor fields, using the Hencky strain and other continuum-mechanical measures. Applications include mapping patterns of brain growth in children, and detecting drug effects on the brain. We are also developing new methods for the registration, segmentation, and analysis of DTI data (diffusion tensor images; shown in the right column here), using Riemannian manifold methods, tensor denoising, and information theory. One project investigates genetic influences on fiber architecture in the brain, using DTI data from 1150 twins. [This is joint work with Ming-Chang Chiang, Alex Leow, Natasha Lepore, Caroline Brun, Yi-Yu Chou, Agatha Lee, and other members of our team].
DIFFUSION TENSOR IMAGING (and HARDI, ULTRA-HIGH FIELD DTI)
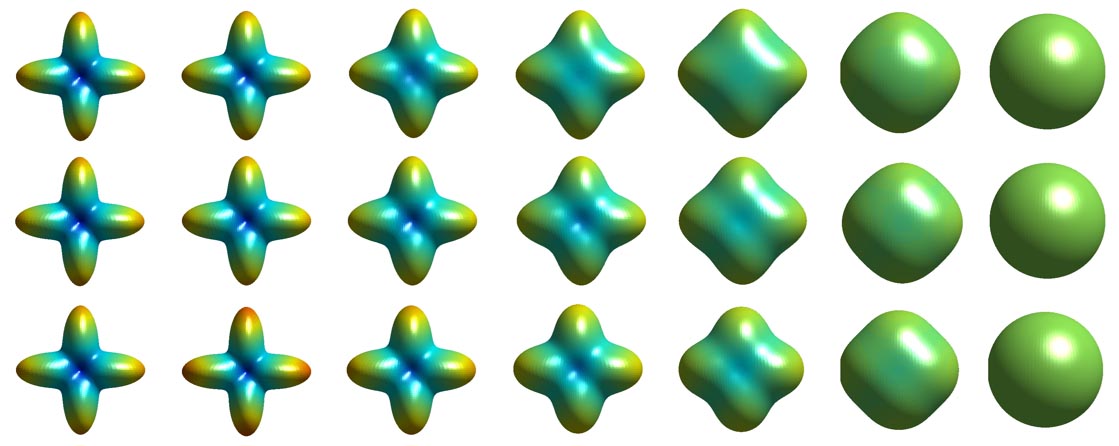

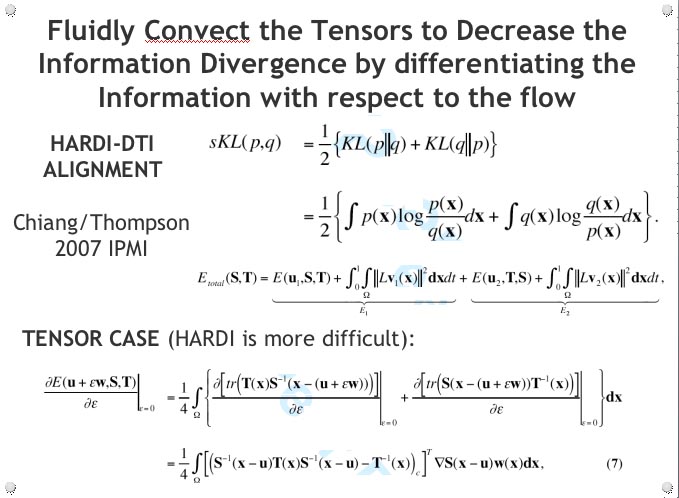
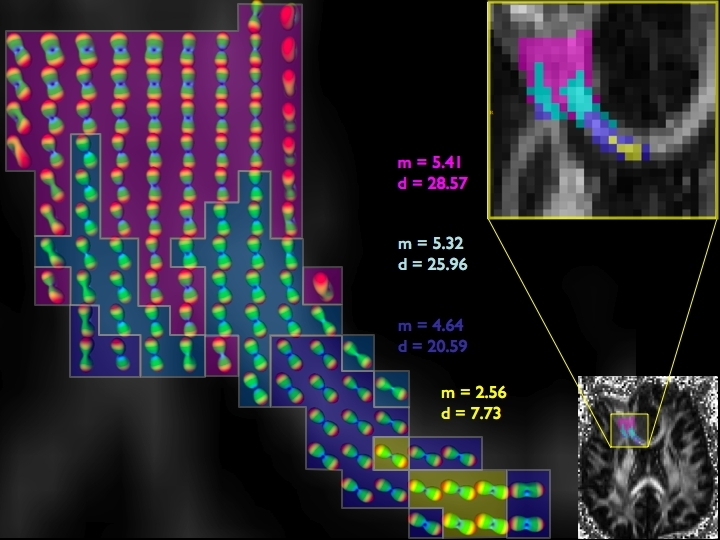

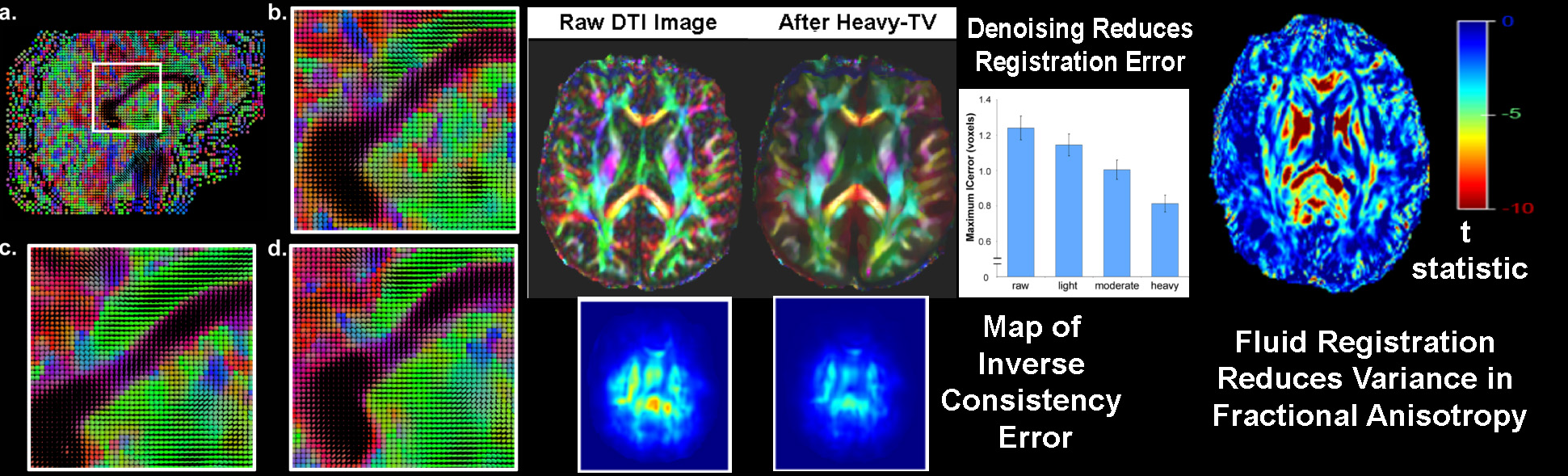
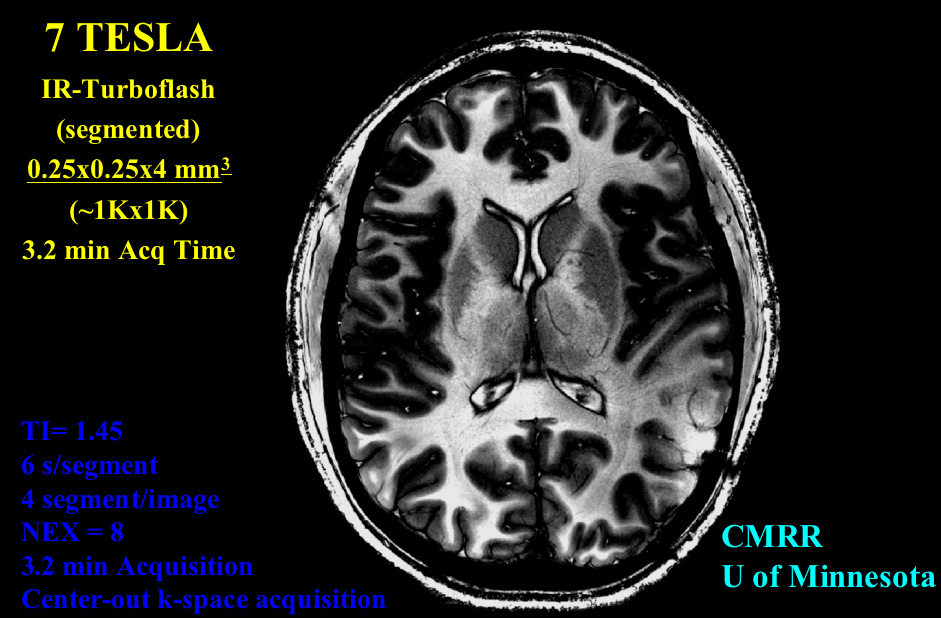
COMPUTER VISION/PATTERN RECOGNITION - AUTOMATED IDENTIFICATION AND LABELING OF BRAIN STRUCTURES
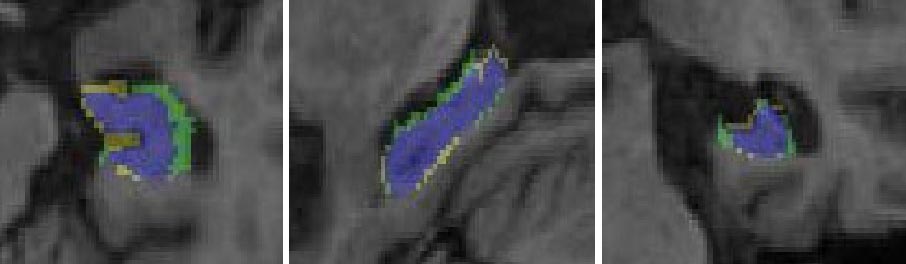
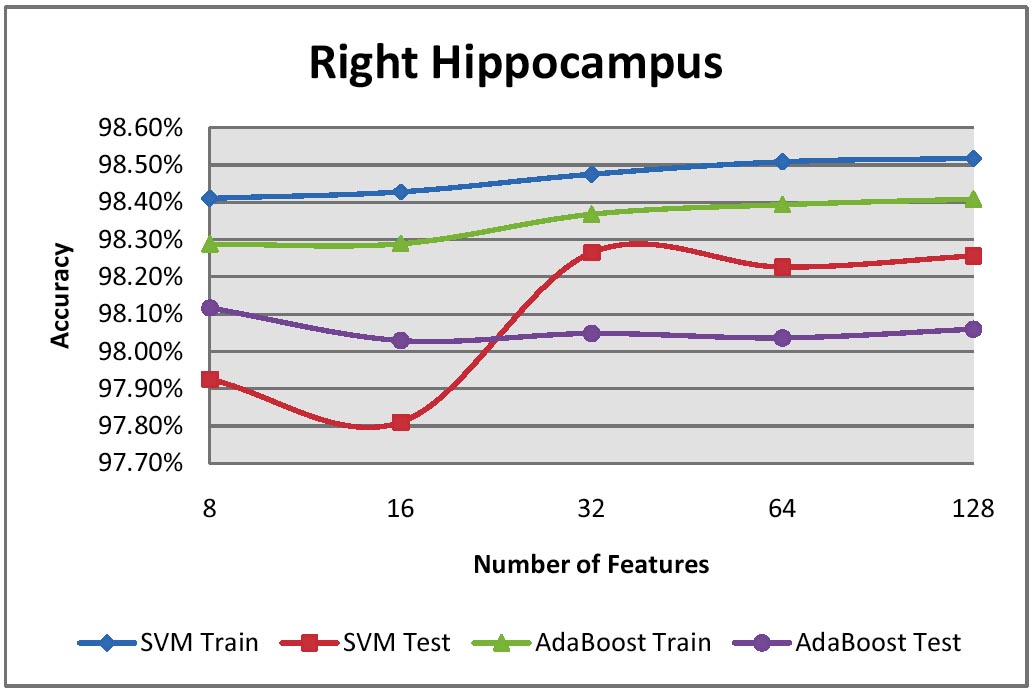
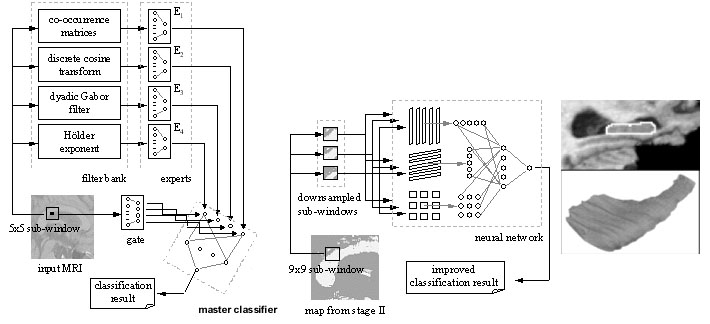
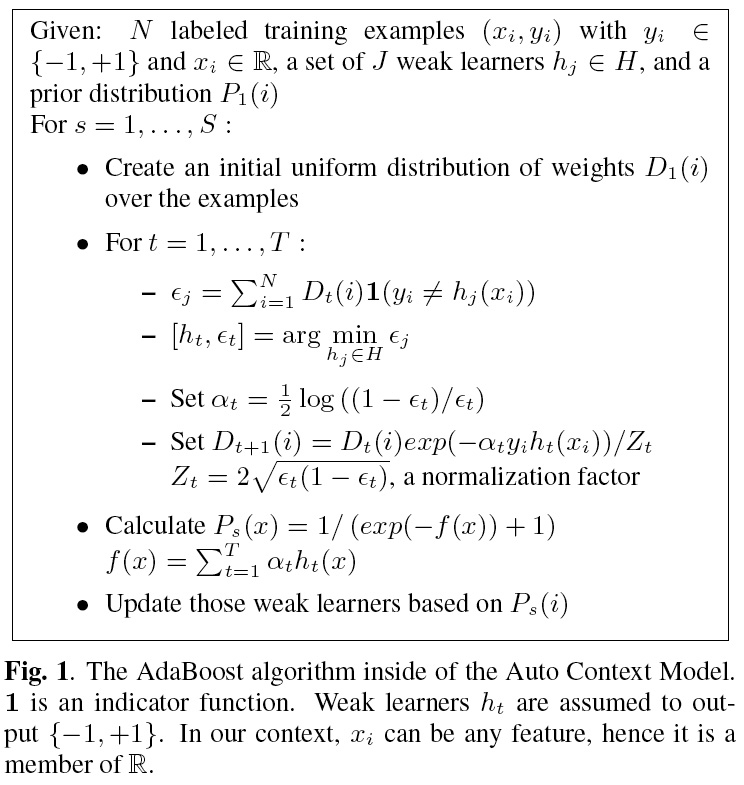
 We have two very active projects that develop pattern recognition and machine learning
methods to automatically find structures
in brain images. Because this is automated, thousands of images can be analyzed rapidly.
Our Ada-SVM project combines Adaptive Boosting and Support Vector Machines, two powerful
computer vision approaches, to automatically extract models of brain structures. We have
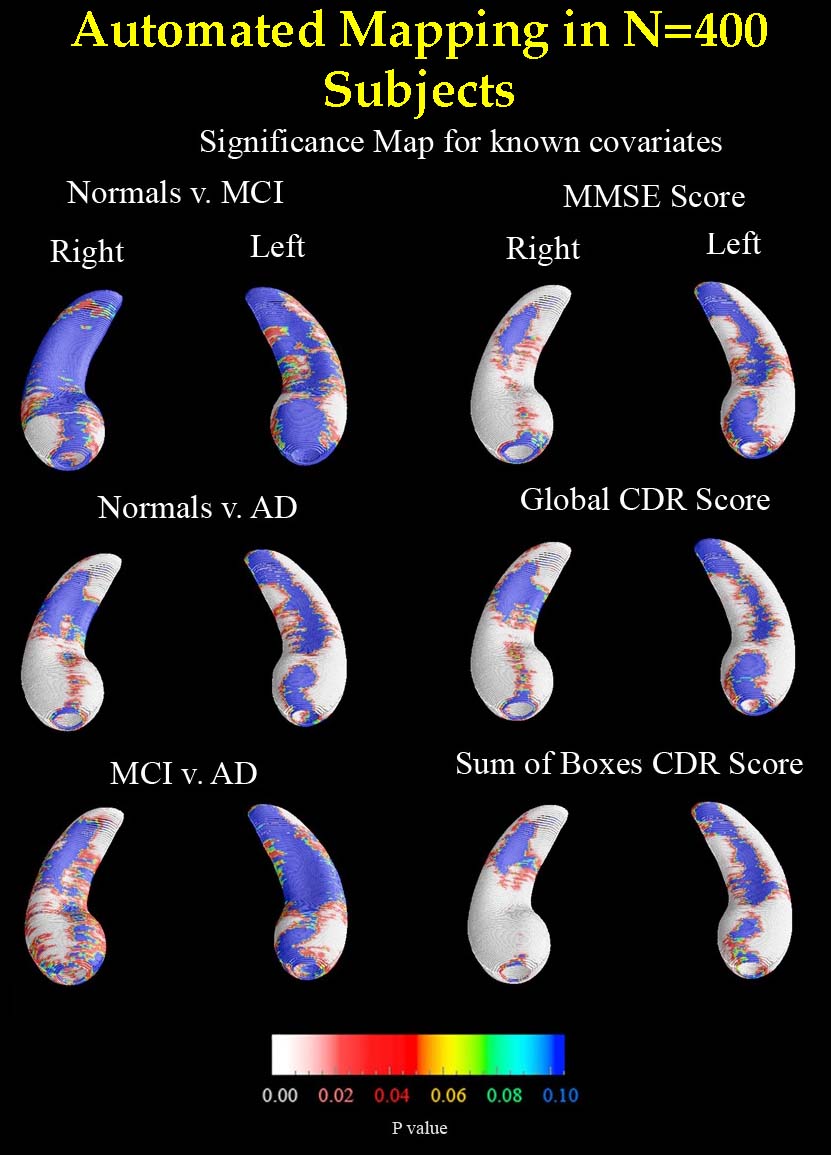
applied this to map factors that affect brain degeneration in over 400 subjects' scans (see figure). Our MAFIA
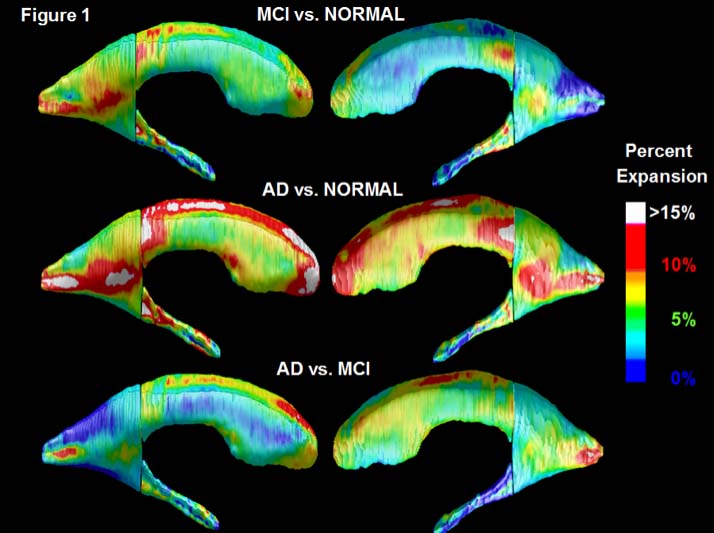
project develops an approach known as Multi-Atlas Fluid Image Alignment (or MAFIA). Here large numbers of surface
models are fluidly deformed, using a Navier-Stokes or Riemannian fluid registration model, onto the new 3D brain scan that needs
to be labeled, and these estimates of the labeled structures are combined using a meta-algorithm to create detailed shape models of
different structures in the brain, such as the lateral ventricles. Vast numbers of images can be
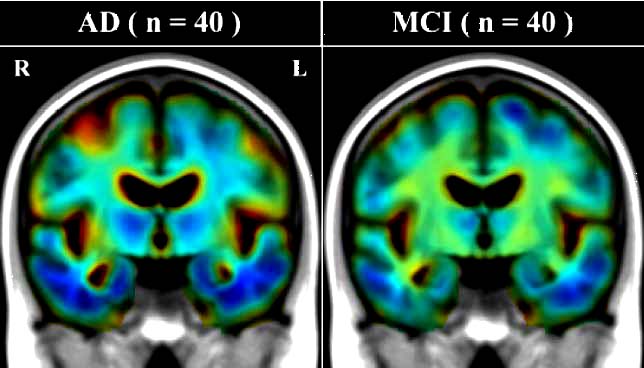
efficiently analyzed, e.g., in drug trial to determine if a treatment slows brain disease progression (see
Figure for automatically computed comparisons of Alzheimer's, mild cognitive impairment, and normal aging). [This is joint work with Jon Morra
and Zhuowen Tu, Yi-Yu Chou, Natasha Lepore, and many others].
We have two very active projects that develop pattern recognition and machine learning
methods to automatically find structures
in brain images. Because this is automated, thousands of images can be analyzed rapidly.
Our Ada-SVM project combines Adaptive Boosting and Support Vector Machines, two powerful
computer vision approaches, to automatically extract models of brain structures. We have
applied this to map factors that affect brain degeneration in over 400 subjects' scans (see figure). Our MAFIA
project develops an approach known as Multi-Atlas Fluid Image Alignment (or MAFIA). Here large numbers of surface
models are fluidly deformed, using a Navier-Stokes or Riemannian fluid registration model, onto the new 3D brain scan that needs
to be labeled, and these estimates of the labeled structures are combined using a meta-algorithm to create detailed shape models of
different structures in the brain, such as the lateral ventricles. Vast numbers of images can be
efficiently analyzed, e.g., in drug trial to determine if a treatment slows brain disease progression (see
Figure for automatically computed comparisons of Alzheimer's, mild cognitive impairment, and normal aging). [This is joint work with Jon Morra
and Zhuowen Tu, Yi-Yu Chou, Natasha Lepore, and many others].
| UNDERSTANDING ANATOMICAL VARIABILITY |
|---|






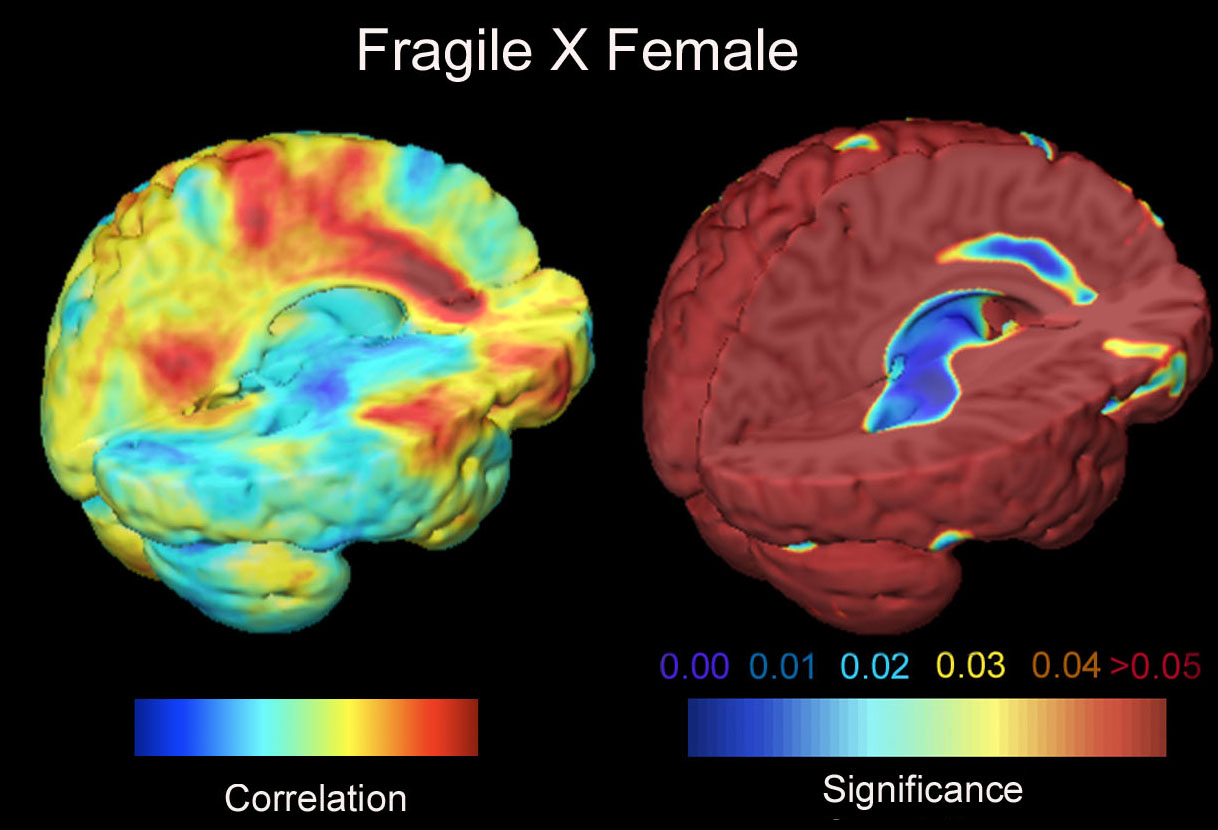
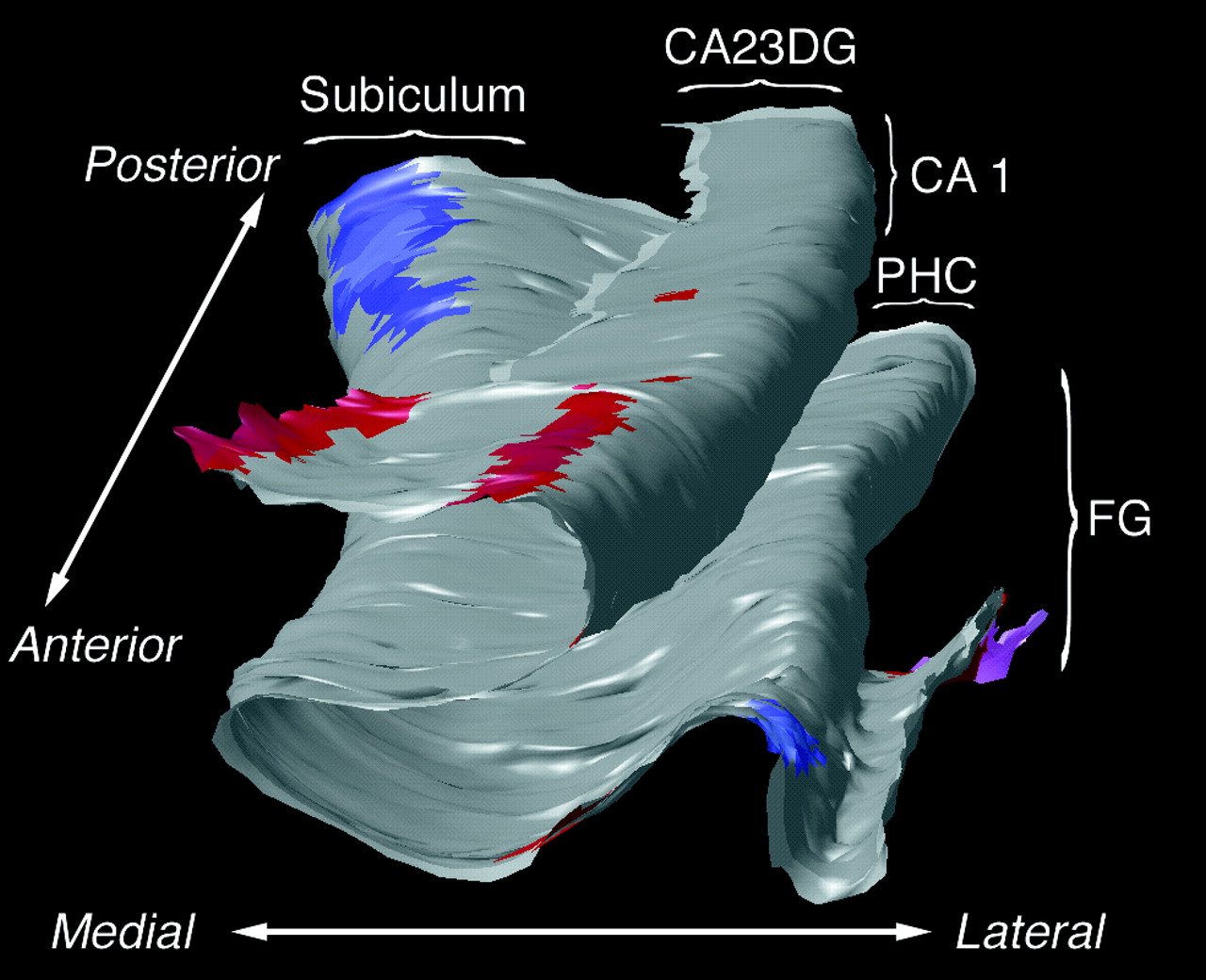
We are interested in modeling the extremely complex variations in brain structure across different human subjects. Here curved landmarks on the brain's cerebral cortex are identified and their variability is represented using covariance tensors. We use this information to detect abnormalities in brain structure. We use covariant partial differential equaltions (PDEs), Green's function methods and random field theory for modeling the geometric variability of the brain. A new direction in this work estimates 6-dimensional Lambda-Tensors, which describe the degree to which variation in one brain area predicts variation in another area of the brain (as a 6x6 tensor for each pair of points in the brain). These tensors can detect regularities and abnormalities in the brain that would otherwise be missed. We also use fractal analysis to understand brain surface complexity. [This is joint work between our team and Pierre Fillard, Vincent Arsigny, Xavier Pennec, and Nicholas Ayache, and others; we have an active exchange program grant to exchange researchers in collaborations between our team at UCLA and the INRIA-Asclepios group in Sophia-Antipolis, near Nice, in the south of France.]
| SURFACE PARAMETERIZATION AND MATCHING |
|---|






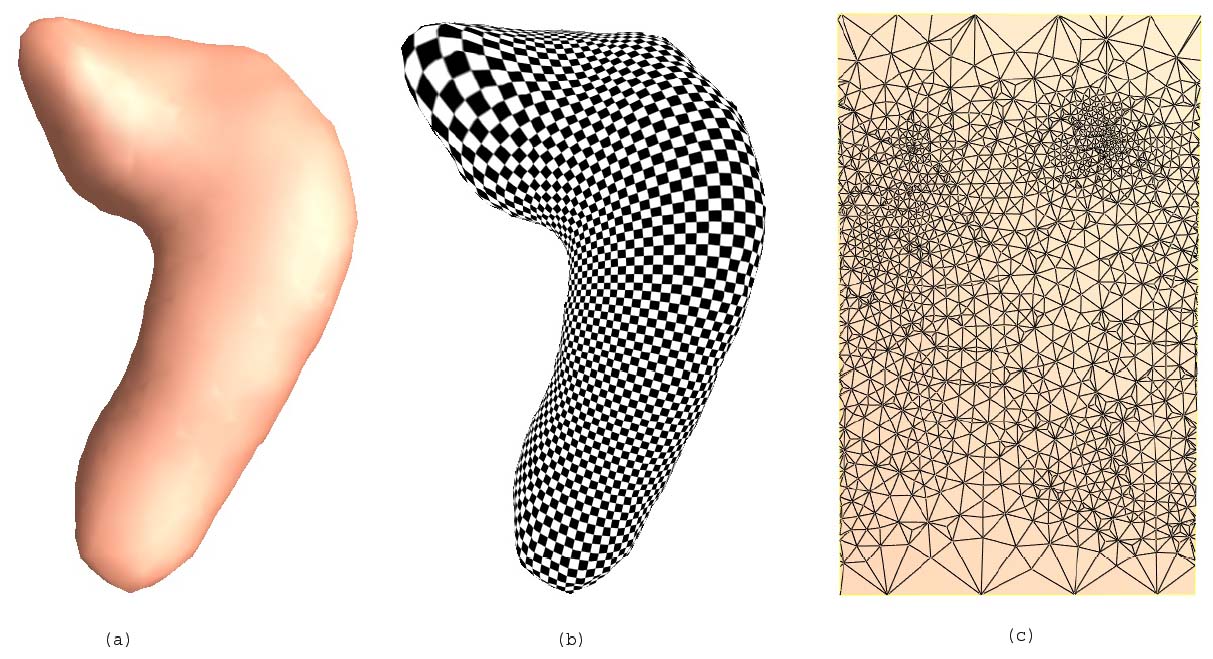
We also work on imposing regular grids on 3D anatomical surfaces in the brain, a process known as surface parameterization. This helps us perform computations on surfaces, such as calculating deformations that match one 3D brain surface with another. We use differential geometry methods such as exterior calculus (differential 1-forms), homology theory, surface-based mutual information, Ricci and Yamabe flows, and p-harmonic mapping methods to analyze features on surfaces. We also use high-dimensional diffeomorphic mappings, computed with spectral methods (i.e. eigenfunctions of self-adjoint operators), to compute flows that align features on surfaces. We also represent these features as level sets (i.e. implicit functions) to help compute mappings between surfaces. These approaches are useful for comparing one brain with another, and for developing statistics on normal and abnormal brain structure and function. [This is joint work with Yalin Wang, Shing-Tung Yau, Alex Leow, Ming-Chang Chiang, Henry Huang, Natasha Lepore, Yonggang Shi, Stanley Osher, Guillermo Sapiro, and others]
| IF YOU'D LIKE MORE INFORMATION... |
|---|
| PUBLICATIONS |CONTACT ME |RESUME |HOMEPAGE |
|---|