 video
video
|
PAUL THOMPSON'S
CURRENT NEUROSCIENCE PROJECTS
(FOR MATH PROJECTS click here) |
|---|
 symbols below).
Contact me if you have any questions!
If you are a mathematician or an engineer, you
may prefer to look at these projects.
symbols below).
Contact me if you have any questions!
If you are a mathematician or an engineer, you
may prefer to look at these projects.




| BRAIN MAPPING IN LARGE HUMAN POPULATIONS |
|---|
Patient populations being studied include large numbers of subjects with Alzheimer's Disease, with mild cognitive impairment, and people at genetic risk for Alzheimer's Disease. Another project studies how HIV/AIDS damages the brain. Other active projects focus on understanding brain changes and drug effects in schizophrenia and bipolar disorder. We are studying chronically-medicated populations, first-episode patients, twins discordant for schizophrenia, and children and adolescents with early onset schizophrenia. Our interest in how schizophrenia develops has also led us to expand our studies of brain development . We also study bipolar disorder (in adolescents and adults) and drug effects on the brain (lithium, antipsychotics, anti-dementia drugs). We are creating new approaches to map how the brain grows in childhood and in the teenage years. Finally, we are especially interested in mapping genetic influences on brain structure. Even in normal individuals, it is intriguing to understand how our genes (and other factors) affect our brain structure and function. It can also help us investigate the genetic causes and inherited risks for disease. Part of this work involves constructing population-based brain atlases to encode and represent patterns of anatomic variation, and to detect structural differences in health and disease. These approaches often use some very interesting mathematics as well as high-performance computing techniques. We also have active research projects on methamphetamine abuse, autism, Williams syndrome, fragile X syndrome, brain asymmetry, and IQ (cognitive ability).
| NORMAL AGING & ALZHEIMER'S DISEASE |
|---|
 In one project, we are creating the first time-lapse maps of Alzheimer's disease spreading in the living brain.
We are building a population-based atlas of the
brain in Alzheimer's Disease. This is a
framework for analyzing brain
data in Alzheimer's patients. It combines images and 3D anatomical models with
MRI, PET, SPECT and histologic data from large numbers of patients with dementia.
Our goal is to determine
3D maps of structural
differences, patterns of gray matter loss, anatomic variability, brain asymmetry,
and regional atrophy
in populations of normal elderly subjects, patients with mild cognitive impairment (MCI),
and patients diagnosed with, or at genetic risk for, Alzheimer's Disease. In these projects,
new mathematical and computational
approaches are being developed, tested and used for analyzing MRI-derived
structural models of the cortex, deep sulci, ventricular system, basal ganglia,
corpus callosum, amygdala and hippocampal formation.
We are especially interested in
understanding the patterns of degenerative rates over time. In an MCI clinical trial, we are investigating
how these profiles of degeneration
can be detected early and slowed down by therapy. We are also developing imaging strategies for
early detection and mapping of degenerative change. A new project is linking cortical thinning in Alzheimer's with amyloid burden
measured in living patients using FDDNP-positron emission tomography.
In one project, we are creating the first time-lapse maps of Alzheimer's disease spreading in the living brain.
We are building a population-based atlas of the
brain in Alzheimer's Disease. This is a
framework for analyzing brain
data in Alzheimer's patients. It combines images and 3D anatomical models with
MRI, PET, SPECT and histologic data from large numbers of patients with dementia.
Our goal is to determine
3D maps of structural
differences, patterns of gray matter loss, anatomic variability, brain asymmetry,
and regional atrophy
in populations of normal elderly subjects, patients with mild cognitive impairment (MCI),
and patients diagnosed with, or at genetic risk for, Alzheimer's Disease. In these projects,
new mathematical and computational
approaches are being developed, tested and used for analyzing MRI-derived
structural models of the cortex, deep sulci, ventricular system, basal ganglia,
corpus callosum, amygdala and hippocampal formation.
We are especially interested in
understanding the patterns of degenerative rates over time. In an MCI clinical trial, we are investigating
how these profiles of degeneration
can be detected early and slowed down by therapy. We are also developing imaging strategies for
early detection and mapping of degenerative change. A new project is linking cortical thinning in Alzheimer's with amyloid burden
measured in living patients using FDDNP-positron emission tomography.
SCHIZOPHRENIA:
MAPPING
BRAIN
CHANGES AND
MEDICATION
EFFECTS
 video
video |
|---|
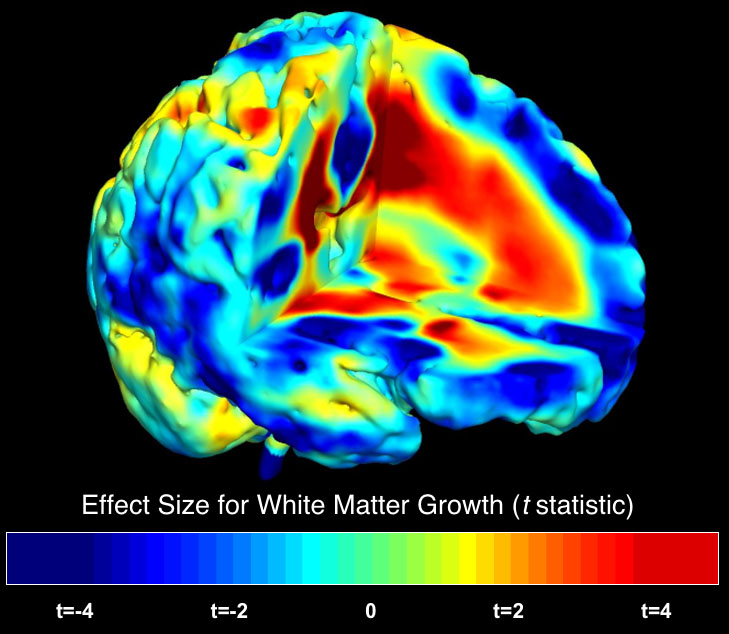
Several exciting studies of schizophrenia are underway. Using novel brain mapping
approaches that we developed for tracking subtle changes in the brain over time,
we are analyzing chronically medicated patients, neuroleptic naive patients, twins discordant for schizophrenia,
and children and adolescents with early-onset schizophrenia. We recently charted
a dynamic wave of gray matter
loss that occurs as schizophrenia develops.
We are interested to see if this implicates some
non-genetic trigger
in
initiating the disease.
In a clinical trial, we are investigating how antipsychotic drugs
such as haloperidol and olanzapine decelerate these brain changes. Using functional MRI (fMRI), we are also studying
patterns of brain activation in
schizophrenia,
and how they relate to genetic polymorphisms and underlying changes in cortical structure.
|
4D
BRAIN
MAPPING:
ANALYZING
GROWTH
PATTERNS
IN NORMAL BRAIN DEVELOPMENT, AUTISM, WILLIAMS SYNDROME, AND FRAGILE X SYNDROME |
|---|
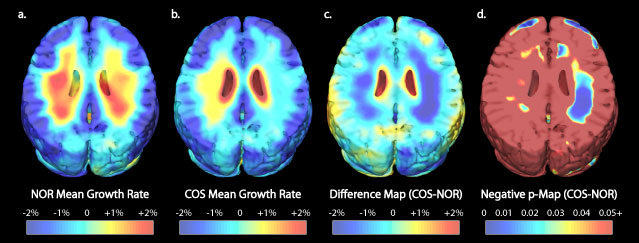
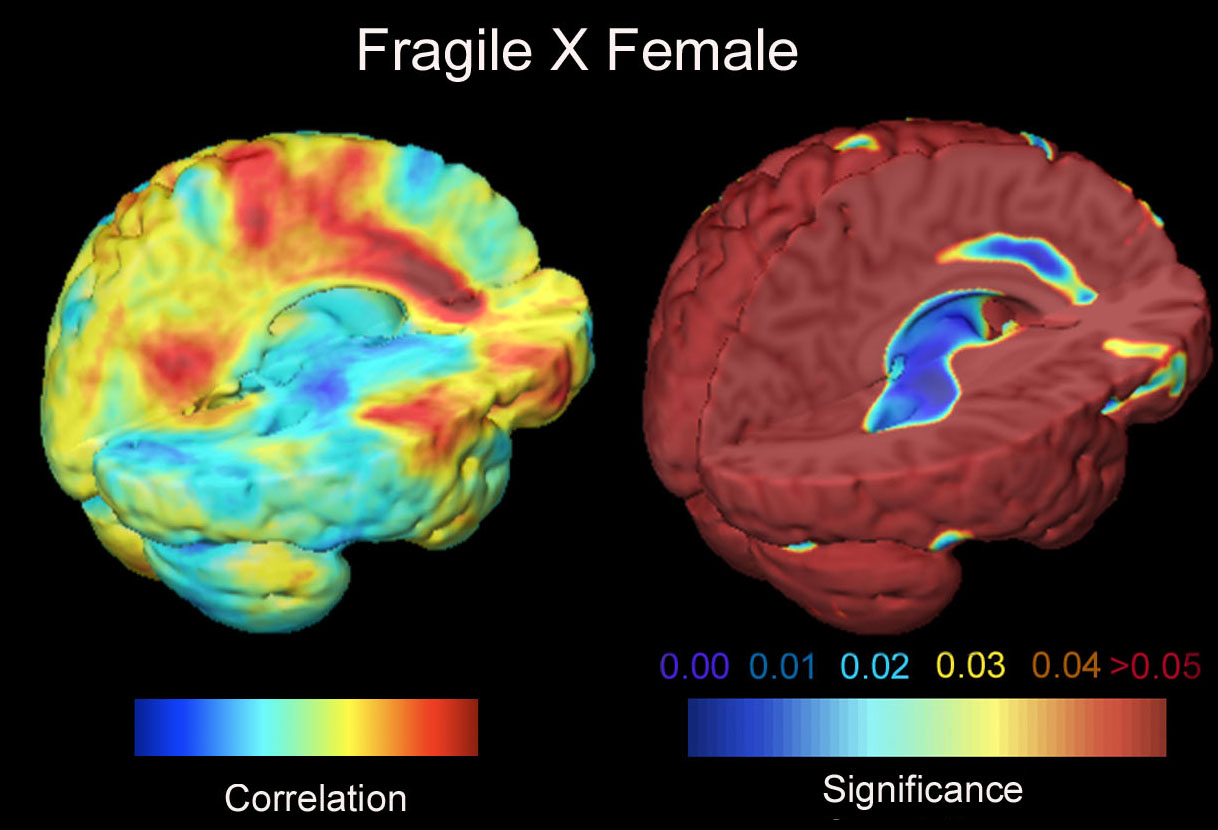 In another project, we are analyzing growth patterns during normal and
abnormal brain development. There are some exciting findings, which we reported recently
in the journal Nature, exploring
the
extremely complex dynamics of
brain growth in children ranging from
6 to 15 years of age. Some very nice articles were written about these
findings in the news media, and these can be found here. Detailed maps
of growth patterns in young children, who were scanned repeatedly with MRI,
can be found here. Current work is creating
time-lapse movies of brain development based on
repeated MRI scans of children as they grow up. We are studying how these brain changes are
accelerated or derailed in children and teenagers
who develop schizophrenia,
or bipolar disorder. We are also developing new brain mapping techniques to help study brain changes in
developmental disorders. We are mapping cortical and subcortical structural differences in
autistic children and Fragile X syndrome. Another project maps subtle cortical
abnormalities in a
genetic disorder of brain development: Williams syndrome, where we
recently mapped the profile of cortical abnormalities.
In another project, we are analyzing growth patterns during normal and
abnormal brain development. There are some exciting findings, which we reported recently
in the journal Nature, exploring
the
extremely complex dynamics of
brain growth in children ranging from
6 to 15 years of age. Some very nice articles were written about these
findings in the news media, and these can be found here. Detailed maps
of growth patterns in young children, who were scanned repeatedly with MRI,
can be found here. Current work is creating
time-lapse movies of brain development based on
repeated MRI scans of children as they grow up. We are studying how these brain changes are
accelerated or derailed in children and teenagers
who develop schizophrenia,
or bipolar disorder. We are also developing new brain mapping techniques to help study brain changes in
developmental disorders. We are mapping cortical and subcortical structural differences in
autistic children and Fragile X syndrome. Another project maps subtle cortical
abnormalities in a
genetic disorder of brain development: Williams syndrome, where we
recently mapped the profile of cortical abnormalities.
| HIV/AIDS EFFECTS ON THE BRAIN |
|---|
 This project studies the damage caused by the HIV virus in the brain. We are creating maps of the regions affected, and assessing
how these
changes relate to cognitive and immune changes in people who are HIV-positive or have AIDS. We are examining how
anti-retroviral treatment influences the rate of brain degeneration in HIV patients. Specific projects focus on the
cerebellum and examining cognitive correlates of progressive brain changes.
This project studies the damage caused by the HIV virus in the brain. We are creating maps of the regions affected, and assessing
how these
changes relate to cognitive and immune changes in people who are HIV-positive or have AIDS. We are examining how
anti-retroviral treatment influences the rate of brain degeneration in HIV patients. Specific projects focus on the
cerebellum and examining cognitive correlates of progressive brain changes.
| METHAMPHETAMINE EFFECTS ON BRAIN STRUCTURE |
|---|
| TUMOR GROWTH: MAPPING RESPONSE TO THERAPY |
|---|
GENETIC
INFLUENCES ON
BRAIN
STRUCTURE
 video
video |
|---|
 We have a major funded effort to determine
how our
genes affect brain structure, function and fiber connectivity.
Genetic brain maps, in particular,
can show whether we inherit patterns of brain structure from our parents, and if so, to what degree. We especially
want to understand
which parts of the brain are most strongly determined by our genes. We are scanning 700 twins with
high-angular resolution diffusion imaging (HARDI) at 4 Tesla to examine which genes influence fiber
integrity and connectivity in the brain. (Read more about HARDI on the math page.) Parallel studies are designed
to help understand
familial
risk for diseases that affect the brain, as well as their genetic transmission.
We have a major funded effort to determine
how our
genes affect brain structure, function and fiber connectivity.
Genetic brain maps, in particular,
can show whether we inherit patterns of brain structure from our parents, and if so, to what degree. We especially
want to understand
which parts of the brain are most strongly determined by our genes. We are scanning 700 twins with
high-angular resolution diffusion imaging (HARDI) at 4 Tesla to examine which genes influence fiber
integrity and connectivity in the brain. (Read more about HARDI on the math page.) Parallel studies are designed
to help understand
familial
risk for diseases that affect the brain, as well as their genetic transmission.
| IMAGING ALGORITHMS: DEVELOPING NEW IMAGE ANALYSIS APPROACHES |
|---|
| DETECTION OF ABNORMAL BRAIN STRUCTURE |
|---|
Often disease-specific patterns that are hard to see in an individual brain scan begin to emerge
after averaging brain data from large numbers of subjects. Construction of a
population-based brain atlas requires the warping of large numbers of brain
images into structural correspondence, prior to the estimation of an
anisotropic Gaussian random field, to represent the calculated structural variations,
on a high-resolution image lattice.
Given a 3D brain image of a
new subject, the algorithms calculate a set of high-dimensional volumetric maps,
typically with 384x384x256x3 (~0.1 billion) degrees of freedom, fluidly
deforming this
scan into structural correspondence with other normal scans, selected one by
one from an anatomic image archive. The family of volumetric warps so
constructed encodes statistical properties of local anatomical variation
throughout the architecture of the brain. Probability maps
can then be generated, which quantify the severity of structural
abnormalities in the anatomy of the new subject. These new techniques for brain image analysis
are reviewed here, and less mathematically
here. Our development of supercomputing approaches to help solve these problems is reviewed
here.
| IF YOU'D LIKE MORE INFORMATION... |
|---|
| PUBLICATIONS |CONTACT ME |RESUME |HOMEPAGE |
|---|