|
by
Paul Thompson

|
I. Challenges in 3D Human Brain Mapping
The rapid growth in brain imaging technologies has been matched by an extraordinary increase in the number of investigations focusing on the structural and functional organization of the brain. Human brain structure is so complex and variable across subjects that engineering approaches drawn from computer vision, image analysis, computer graphics and artificial intelligence research fields are required to manipulate, analyze and communicate brain data.
Extreme variations in brain structure, especially in the gyral patterns of the human cortex, present two major types of challenges in brain mapping studies. First, anatomic variations make it especially difficult to design computerized strategies for detecting abnormal brain structure. Analysis of regional neuroanatomy and cortical morphology are key factors in the radiologic assessment of a wide range of neurological disorders, including Alzheimer's Disease, Pick's Disease and other dementias (Friedland and Luxenberg, 1988), schizophrenia (Kikinis et al., 1994), epilepsy (Cook et al., 1994), cortical dysplasias (Sobire et al., 1995), and other developmental disorders. To distinguish abnormalities from normal variants, a realistically complex mathematical framework is needed to encode information on anatomic variability in homogeneous populations (Grenander and Miller, 1994). Secondly, integrating and comparing data from multiple subjects and groups is hampered by the extreme complexity of anatomic variations (Meltzer and Frost, 1994; Woods, 1996). Ideally, when analyzing functional imaging data, we would like to remove all morphological differences between individual brains before considering the distribution of functional information on the anatomic substrate. As a result, both the detection of structural abnormalities in disease and the pooling of multi-subject brain data present considerable challenges, both neurobiological and mathematical in nature.
These difficulties have prompted us to explore hybrid approaches for brain image registration and pathology detection. In these approaches, computer vision algorithms and statistical pattern recognition measures are integrated with anatomically-driven elastic transformations which encode complex shape differences between systems of anatomic surfaces. These algorithms can help to integrate brain data from many subjects, and to obtain objective criteria for conditions such as global or regional cerebral atrophy, as well as the gyral and sulcal anomalies specific to certain disease states.
Digital Brain Atlases. To illustrate these challenges, we consider the recent rapid progress made by research groups developing standardized three-dimensional atlases of the human brain (Talairach and Tournoux, 1988; Greitz et al., 1991; Hohne et al., 1992; Thurfjell et al., 1993; Kikinis et al., 1996). Most brain atlases are based on a detailed representation of a single subject's anatomy in a standardized 3D coordinate system, or stereotaxic space. These atlases provide an invariant reference system, and the possibility of template matching, allowing anatomical structures in new scans to be identified and analyzed. Digital overlay of the Schaltenbrand, Talairach and Brodmann atlas data onto volumetric radiologic scans can create composite maps and simulation displays for surgical planning (Hardy, 1994). However, in view of the complex structural variability between individuals, a fixed brain atlas may not serve as a faithful representation of every brain (Roland and Zilles, 1994; Mazziotta et al., 1995). It would therefore be ideal if an atlas could either be (1) elastically deformed to fit a new image set from an incoming subject, or (2) expanded to incorporate neuroanatomic data from large populations of human subjects (Mazziotta et al., 1995).
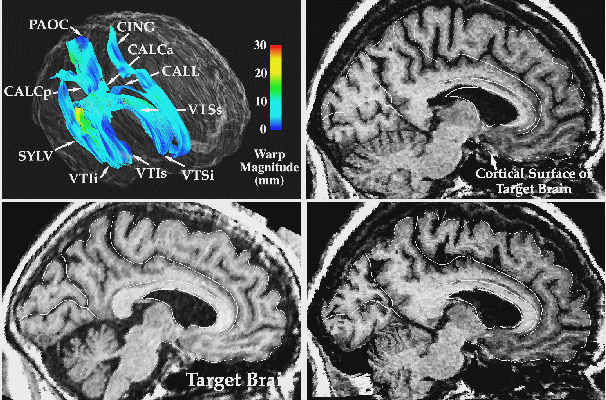
Deformable and Probabilistic Brain Atlases. Deformable atlases are brain atlases which can be adapted to reflect the anatomy of new subjects (Evans et al., 1991; Gee et al., 1993; Christensen et al., 1993; Sandor and Leahy, 1995; Rizzo et al., 1995; Haller et al., 1997). Image warping algorithms, specially designed to handle 3D neuroanatomic data, apply complex profiles of dilation and contraction to a digital brain atlas, reconfiguring it into the shape of the patient's anatomy. Any successful warping transform for individualizing a brain atlas must be high-dimensional, i.e. it must allow any segment of the atlas anatomy to dilate, contract, twist and even rotate, to bring the atlas into structural correspondence with the target scan at a very local level (Christensen et al., 1996). High-dimensional brain image registration, or warping algorithms (Christensen et al., 1993; 1996; Collins et al., 1994a; Thirion, 1995; Rabbitt et al., 1995; Warfield et al., 1995; Davatzikos, 1996; Thompson and Toga, 1996c, Bro-Nielsen and Gramkow, 1996) can transfer all of the information in a 3D digital brain atlas onto the scan of any given subject, while respecting the intricate patterns of structural variation in their anatomy. Such deformable atlases can carry 3D maps of functional and vascular territories into the coordinate system of different patients, as well as information on different tissue types and the boundaries of cytoarchitectonic fields and their neurochemical composition. In addition, the complex profiles of dilation and contraction required to warp an atlas onto the new subject's brain provide an index of the anatomical shape differences between that subject's brain and the atlas. Differences in regional shape can be assessed by applying vector and tensor field operators to the transformation field required to locally deform one brain volume into another (Thompson et al., 1998; Thirion et al., 1998). Pathology detection algorithms will also be discussed later. These invoke deformation fields which match one brain with a large number of others. The result is a probabilistic brain atlas which encodes patterns of anatomic variation in human populations, and incorporates algorithms to detect structural variants outside of the normal range (Mazziotta et al., 1995; Thompson et al., 1997).
Brain to Atlas Transformations. In functional imaging, neuroanatomic atlas data acts as a template on which other brain maps can be overlaid. As well as providing structural perspectives such as chemoarchitecture, the atlas provides the additional detail necessary to accurately localize activation sites. Digital mapping of structural and functional image data into a common 3D coordinate space is a prerequisite for many types of brain imaging research, as it supplies a quantitative spatial reference system in which brain data from multiple subjects and modalities can be compared and correlated. To bring new brain data into register with an atlas template, warping algorithms are vital to compensate for morphologic differences among individual brains. These differences can otherwise hinder the pooling and comparison of multi-subject brain data. Transforming individual datasets into the shape of a single reference anatomy, or onto a 3D digital brain atlas, removes subject-specific shape variations, and allows subsequent comparison of brain function between individuals (Christensen et al., 1993; Ashburner et al., 1997). Warping transforms can also correct for complex structural change due to histologic processing, allowing direct correlation of post mortem neurochemical maps with functional or metabolic image data acquired from the same subject in vivo (Mega et al., 1997).
Measuring Brain Changes. Finally, when applied to scans acquired over many months or years from a single subject, 3D warping algorithms can calculate measures of local and global shape change over time (Toga et al., 1996; Thompson et al., 1998). In many ways, static representations of brain structure are ill-suited to investigating dynamic processes of disease. With warping algorithms, measures of dilation rates, contraction rates, and rates of shearing and divergence of the cellular architecture may be computed locally, for all structures, directly from the deformation field which matches one scan with the other. As a result, warping algorithms account for the anatomic variations and idiosyncrasies of individual subjects, and offer a powerful strategy to track temporal change and classify age-related, developmental or pathologic alterations in anatomy.
II. Classification of Warping Algorithms
Model-Driven and Intensity-Driven Algorithms. A wide variety of 3D image warping algorithms have been designed to handle neuroanatomic data. They can, however, be classified according to their salient characteristics. Model-driven algorithms first build explicit geometric models, representing separate, identifiable anatomic elements in each of the scans to be matched. These anatomical systems typically include functionally important surfaces (Szeliski and Lavallee, 1993; Downs et al., 1994; Moshfeghi et al., 1995; Thompson and Toga, 1996; Davatzikos, 1996), curves (Ge et al., 1995; Monga and Benayoun, 1995; Subsol, 1995), and point landmarks (Bookstein, 1989; Amit et al., 1991). Anatomical elements are each parameterized and matched with their counterparts in the target scan, and their correspondences guide the volumetric transformation of one brain to another. In our own warping algorithm (Section 3; Thompson and Toga, 1996, 1998), higher-level structural information guides the mapping of one brain onto another, and a hierarchy of curve-to-curve and surface-to-surface mappings is set up, guaranteeing the biological validity of the resulting transform. Specialized approaches are also developed which exploit anatomical information to match cortical regions, so that networks of sulci and gyri are individually matched. These strategies are discussed in Section 3.
Model-driven approaches contrast with intensity-driven approaches. Intensity-driven approaches aim to match regional intensity patterns in each scan based on mathematical or statistical criteria. Typically, they define a mathematical measure of intensity similarity between the deforming scan and the target. Measures of intensity similarity can include squared differences in pixel intensities (Christensen et al., 1993), regional correlation (Bajcsy and Kovacic, 1989), or mutual information (Kim et al., 1997). Mutual information has proved to be an excellent similarity measure for cross-modality registrations, since it assumes only that the statistical dependence of the voxel intensities is maximal when the images are geometrically aligned. The intensity similarity measure, combined with a measure of the structural integrity of the deforming scan, is optimized by adjusting parameters of the deformation field. Algorithms based on intensity patterns alone essentially by-pass information on the internal topology of the brain. Instead, mathematical criteria are used in an attempt to automatically discriminate among a large number of potential false matches in each dataset. Matching of neuroanatomic data in the absence of higher-level structural information presents an extremely difficult pattern recognition problem. Future hybrid approaches, based on a combination of model-based and densitometric criteria, are likely to benefit from the advantages of each strategy.
Intensity-Driven Approaches
A key insight, which spurred the development of intensity-based warping algorithms, was the connection of the image data with a physically deforming system in 3 dimensions (Broit, 1981). Physical continuum models consider the deforming image to be embedded in a 3-dimensional deformable medium, which can be either an elastic material or a viscous fluid. The medium is subjected to certain distributed internal forces, which reconfigure the medium and eventually lead the image to match the target. These forces can be based mathematically on the local intensity patterns in the datasets, with local forces designed to match image regions of similar intensity.
Navier-Stokes Equilibrium Equations. In elastic media, the displacement field u(x) resulting from internal deformation forces F(x) (called 'body forces') obeys the Navier-Stokes equilibrium equations for linear elasticity:
Here R is a discrete lattice representation of the scan to be transformed,
L is the
Cauchy-Navier operator ![]() , and Lame's coefficients lambda and mu refer to the elastic
properties of
the medium. Body forces, designed to match regions in each dataset with high
intensity similarity, can be derived from the gradient of a local
correlation function. In (Bajcsy and Kovacic, 1989), intensity neighborhoods
to be correlated in each scan were projected onto a truncated 3D Hermite
polynomial basis to enhance the response of edge features and accelerate
computation. More complex local operators can also be designed to identify
candidates for regional matches in the target image (Amit, 1997). With
proper boundary conditions the elasticity equilibrium equations can be
solved numerically by finite difference, finite element, or spectral
methods. This elasticity-based warping scheme was introduced by
Broit (1981). It was subsequently extended to a multiresolution/multigrid
algorithm by Bajcsy and Kovacic (1989), and to a finite element
implementation by Gee et al. (1993).
, and Lame's coefficients lambda and mu refer to the elastic
properties of
the medium. Body forces, designed to match regions in each dataset with high
intensity similarity, can be derived from the gradient of a local
correlation function. In (Bajcsy and Kovacic, 1989), intensity neighborhoods
to be correlated in each scan were projected onto a truncated 3D Hermite
polynomial basis to enhance the response of edge features and accelerate
computation. More complex local operators can also be designed to identify
candidates for regional matches in the target image (Amit, 1997). With
proper boundary conditions the elasticity equilibrium equations can be
solved numerically by finite difference, finite element, or spectral
methods. This elasticity-based warping scheme was introduced by
Broit (1981). It was subsequently extended to a multiresolution/multigrid
algorithm by Bajcsy and Kovacic (1989), and to a finite element
implementation by Gee et al. (1993).
Viscous Fluid Approaches. More recently, Christensen et al. (1993, 1995, 1996) proposed a viscous-fluid based warping transform, motivated by capturing non-linear topological behavior and large image deformations. Designed to operate sequentially, this transform is actually a series of three algorithms which adjust successively finer features of the local anatomy until the transformed image matches the target scan in exquisite detail. The optimal deformation field is defined as the one that maximizes a global intensity similarity function (defined on the deformed template and the target), while satisfying additional continuum-mechanical constraints which guarantee the topological integrity of the deformed template.
The transformation proposed by Christensen et al. (1996) is conducted in 3 successive stages. Stage 1 requires a sparse manual specification of the displacement field by isolating several corresponding landmarks in both 3D scans. The minimum energy configuration of the template compatible with this initial assignment is formalized as a Dirichlet problem (Joshi et al., 1995). For a system of point landmarks, the associated Fredholm integral equation reduces to a linear system whose solution expresses the Stage 1 deformation field in terms of the self-adjoint linear operator describing the mechanics of the deforming system. A second step expresses the residual deformation in terms of an approximation series of eigenfunctions of the linear elastic operator. Basis coefficients are determined by gradient descent on a cost functional (2) which penalizes squared intensity mismatch between the deforming template T(x-u(x,t)) and target S(x):
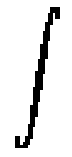 ||
T(x-u(x,t))-S(x)||2
dx (2)
||
T(x-u(x,t))-S(x)||2
dx (2)
Finally, a third, viscous deformation stage allows large-distance non-linear fluid evolution of the neuroanatomic template. With the introduction of concepts such as deformation velocity and a Eulerian reference frame, the energetics of the deformed medium are hypothesized to be relaxed in a highly viscous fluid. The driving force, which deforms the anatomic template, is critical for successful registration, and is defined as the variation of the cost functional with respect to the displacement field.
The deformation velocity is governed by the creeping flow momentum equation for a Newtonian fluid and the conventional displacement field in a Lagrangian reference system is connected to a Eulerian velocity field by the relation of material differentiation. Experimental results were excellent (Christensen et al., 1996).
Convolution Filters. Linkage of continuum-mechanical models with an
3D intensity matching optimization problem results in an extremely
computationally intensive algorithm. Registration of two 128x128x148 MR
volumes took 9.5 and 13 hours for elastic and fluid transforms,
respectively, on a 128x64 DECmpp1200Sx/Model 200 MASPAR (Massively-
Parallel
Mesh-Connected Supercomputer). This spurred work to accelerate or modify the
algorithm for use on standard single-processor workstations (Thirion, 1995;
Bro-Nielsen and Gramkow, 1996). Both elastic and fluid algorithms contain
core systems of up to 0.1 billion simultaneous partial differential
equations, requiring many iterations of successive over-relaxation to
find their solution. To avoid this need to pass a filter many times over the
vector arrays, Bro-Nielsen and Gramkow (1996) developed a convolution filter
to solve the system of partial differential equations in a single pass over
the data. This speeds up the core step in the registration procedure by a
factor of 1000. Since the behavior of the mechanical system is governed by
the Navier-Stokes differential operator L = ![]() , the
eigenfunctions of this operator (Christensen, 1994) were used to derive a
Green's function solution u*(x)=G(x-x0) to the impulse response equation
Lu*(x-x0)=d(x-x0), where d is the Dirac delta-function.
The solution to the full PDE Lu(x)=F(x-u(x)) was
approximated as
a rapid filtering operation on the 3D arrays representing the components of
the body force:
, the
eigenfunctions of this operator (Christensen, 1994) were used to derive a
Green's function solution u*(x)=G(x-x0) to the impulse response equation
Lu*(x-x0)=d(x-x0), where d is the Dirac delta-function.
The solution to the full PDE Lu(x)=F(x-u(x)) was
approximated as
a rapid filtering operation on the 3D arrays representing the components of
the body force:
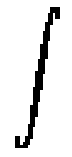 G(x-r).F(r) dr = (G*F)(x),
G(x-r).F(r) dr = (G*F)(x),
where G* represents convolution with the impulse response filter. As noted in (Gramkow and Bro-Nielsen, 1997), a recent fast, demons-based warping algorithm (Thirion, 1995) calculates a flow velocity by regularizing the force field driving the template with a Gaussian filter. Since this filter may be regarded as a separable approximation to the continuum-mechanical filters derived above, interest has focused on deriving additional separable (and therefore rapidly applied) filters to capture the deformational behavior of material continua in image registration (Gramkow, 1996).
Multigrid and Coarse-to-Fine Optimization. Vast numbers of parameters are required to represent complex deformation fields. Optimal values for these parameters must therefore be searched for using robust numerical algorithms, to find parameter sets which globally minimize the measure of mismatch between the warped image and target. In several robust systems for automated brain image segmentation and labeling, Dengler (1988), Bajcsy and Kovacic (1989), Collins (1994a, 1995), and Gee (1993,1995) recover the optimal transformation in a hierarchical multi-scale fashion. Both template and target intensity data are downsampled or smoothed with different sized Gaussian filters, and the registration is performed initially at coarse spatial scales, then finer ones. This accelerates computation and helps to avoid local minima of the mismatch measure. In an alternative approach, the deformation field is expressed as a steadily increasing sum of basis functions, whose coarse-scale parameters are estimated first and whose spatial frequency is increased as the algorithm progresses (Amit et al., 1991; Miller et al., 1993). The widely-used Statistical Parametric Mapping (Ashburner et al., 1997) and Automated Image Registration algorithms (Woods et al., 1998) take a similar coarse-to-fine approach, expressing increasingly complex warping fields in terms of a 3D cosine basis (SPM) or by using 3D polynomials of increasing order (AIR; polynomials also span the space of continuous deformation fields, by the Stone-Weierstrass theorem). Amit (1997) expresses 2D warping fields in terms of a wavelet basis. The small support of high-frequency wavelets allows for local adjustments of the warping field in regions where the mismatch is large, without having to alter the field in other areas where the mismatch may already be small. In (Miller et al., 1993; cf. Amit et al., 1991), a stochastic algorithm generates the expansion coefficient set for the deformation field in terms of an eigenbasis for the linear elasticity operator. Stochastic gradient descent is used to find the optimal warping field parameters. At the expense of added computation time, stochastic sampling allows globally optimal image matches to be estimated.
Bayesian Registration Models. Bayesian statistical models provide a general framework for the presentation of deformable matching methods. Recently, they have also been applied to the brain image matching problem (Miller et al., 1993; Gee et al., 1993, 1995; Ashburner et al., 1997). In a Bayesian model, statistical information on the imaging process (the imaging model) is combined with prior information on expected template deformations (the prior model) to make inferences about the parameters of the deformation field. In fact, all of the intensity-based approaches which combine an intensity mismatch term with a measure of deformation severity can be recast as an inference task in a Bayesian probabilistic framework. The Bayesian maximum a-posteriori (MAP) estimator solving the registration problem is the transformation argmin (D(u)+E(u)) which minimizes a combined penalty due to the intensity mismatch D(u) and the elastic potential energy E(u) stored in the deformation field.
Paul Thompson
| RESUME| E-MAIL ME| PERSONAL HOMEPAGE| PROJECTS |
|---|