
|
Paul M. Thompson and Arthur W. Toga
Laboratory of Neuro Imaging, Department of Neurology, Division of Brain Mapping, UCLA School of Medicine, Los Angeles, California 90095
| REVIEW OF WARPING APPROACHES| CORTICAL WARPING| DEFORMABLE BRAIN ATLASES | RELATED RESEARCH |
|---|


|
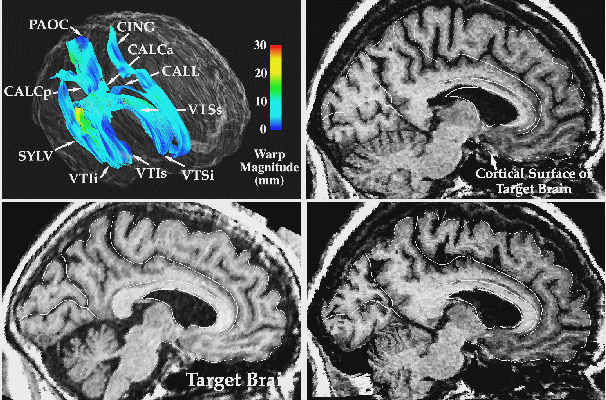
3D Image Warping Measures Patterns of Anatomic Differences. T1-weighted MR sagittal brain slice images from (top left:) a normal elderly subjects scan, (top middle:) a target anatomy, from a patient with clinically-determined Alzheimer's disease; and (bottom left:) result of warping the reference anatomy into structural correspondence with the target. Note the precise non-linear registration of the cortical boundaries, the desired reconfiguration of the major sulci, and the contraction of the ventricular space and cerebellum. Both global and local differences in anatomy are accommodated by the transformation. The complexity of the recovered deformation field is shown by applying the two in-slice components of the 3D volumetric transformation to a regular grid in the reference coordinate system. This visualization technique (bottom, middle panel) highlights the especially large contraction in the cerebellar region, and the complexity of the warping field in the posterior frontal and cingulate areas, corresponding to subtle local variations in anatomy between the two subjects. To monitor the smooth transition to the surrounding anatomy of the deformation fields initially defined on the surface systems, the magnitude of the warping field is visualized on models of the surface anatomy of the target brain, as well as on an orthogonal plane slicing through many of these surfaces at the same level as the anatomic sections (bottom right panel). Note the smooth continuation of the warping field from the complex anatomic surfaces into the surrounding brain architecture, and the highlighting of the severe deformations in the pre-marginal cortex, ventricular and cerebellar areas.
The authors have devised, implemented, and tested a fast, spatially accurate technique for calculating the high-dimensional deformation field relating the brain anatomies of an arbitrary pair of subjects. The resulting three-dimensional (3-D) deformation map can be used to quantify anatomic differences between subjects or within the same subject over time and to transfer functional information between subjects or integrate that information on a single anatomic template. The new procedure is based on developmental processes responsible for variations in normal human anatomy and is applicable to 3-D brain images in general, regardless of modality. Hybrid surface models known as Chen surfaces (based on superquadrics and spherical harmonics) are used to efficiently initialize 3-D active surfaces, and these then extract from both scans the developmentally fundamental surfaces of the ventricles and cortex.
The construction of extremely complex surface deformation maps on the internal cortex is made easier by building a generic surface structure to model it. Connected systems of parametric meshes model several deep sulci whose trajectories represent critical functional boundaries. These sulci are sufficiently extended inside the brain to reflect subtle and distributed variations in neuroanatomy between subjects. The algorithm then calculates the high-dimensional volumetric warp (typically with 384x384x256x3 ~ 0.1 billion degrees of freedom) deforming one 3-D scan into structural correspondence with the other. Integral distortion functions are used to extend the deformation field required to elastically transform nested surfaces to their counterparts in the target scan. The algorithm's accuracy is tested, by warping 3-D magnetic resonance imaging (MRI) volumes from normal subjects and Alzheimer's patients, and by warping full-color 1024x1024x1024 digital cryosection volumes of the human head onto MRI volumes.
Applications are discussed, including the transfer of multisubject 3-D functional, vascular, and histologic maps onto a single anatomic template; the mapping of 3-D brain atlases onto the scans of new subjects; and the rapid detection, quantification, and mapping of local shape changes in 3-D medical images in disease and during normal or abnormal growth and development.
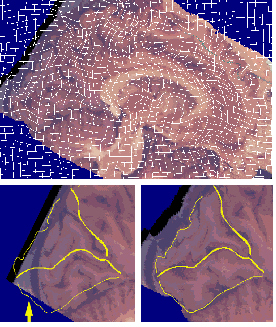
Inter-Modality Warping: Mapping 3D Digital Cryosection Volumes onto 3D MRI Volumes. The result of warping a 3D cryosection image (lower left) into the shape of a target MRI anatomy is shown (lower right), with cortical landmarks of the target anatomy superimposed. Note the reconfiguration of major occipital lobe sulci into the shape of the target anatomy. This type of registration of critical lobar, sulcal and cytoarchitectural boundaries is only possible with a high-dimensional warping technique (cf. Christensen et al., 1996).
|
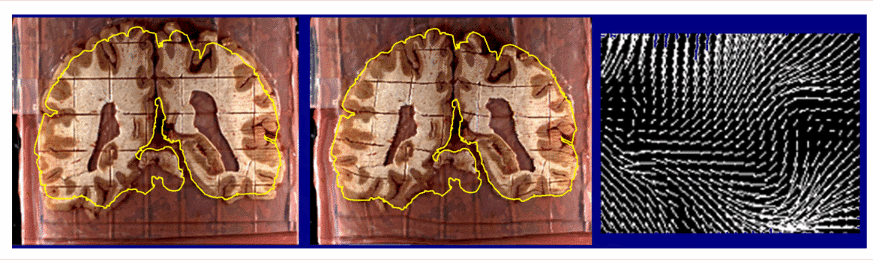
| Recovery of Change in Brain Tissue due to Post Mortem Effects and Histologic Processing. Warping algorithms based on continuum-mechanical models can recover and compensate for patterns of tissue change which occur in post mortem histologic experiments. A brain section (left), gridded to produce tissue elements for biochemical assays. is reconfigured (middle) into its original position in the cryosection blockface (Mega et al., 1997; algorithm from Thompson and Toga, 1996, 1998). The complexity of the required deformation vector field in a small tissue region (magnified vector map, right) demonstrates that very flexible, high-dimensional transformations are essential (Thompson and Toga, 1996; Schormann et al., 1996). As well as measuring local patterns of mechanical tissue deformations, recovery of deformation fields allows projection of histologic and biochemical data back into the volumetric reference space of the cryosection image. In some cases, these data can also be projected, using additional warping algorithms, onto in vivo MRI and co-registered PET data from the same subject for digital correlation and analysis (Mega et al., 1997). |
Paul Thompson
| RESUME| E-MAIL ME| PERSONAL HOMEPAGE| PROJECTS |
|---|