Thompson PM, Mega MS, Woods RP, Blanton RE, Moussai J, Zoumalan CI, Cummings JL, Toga AW
Laboratory of Neuro Imaging, Dept. Neurology, Division of Brain Mapping,
UCLA School of Medicine, Los Angeles CA 90095, USA,
and
UCLA Alzheimer's Disease Center
Summary. We report a new brain atlasing strategy, applied to create the first disease-specific brain atlas. The atlas, constructed to represent the human brain in Alzheimer's Disease (AD), provides a 3-dimensional analysis of structural variation throughout the brain, based on 84 structures per subject. Specialized strategies were developed for accurate group averaging of anatomy, to average cortical topography and to encode its local variation. Disease-specific features and asymmetries emerged cortically and subcortically that were not apparent in the individual anatomies. A sharply-defined mosaic of variability and asymmetry patterns emerged at the cortex, which segregated sharply according to the functional specialization of each system (see below).

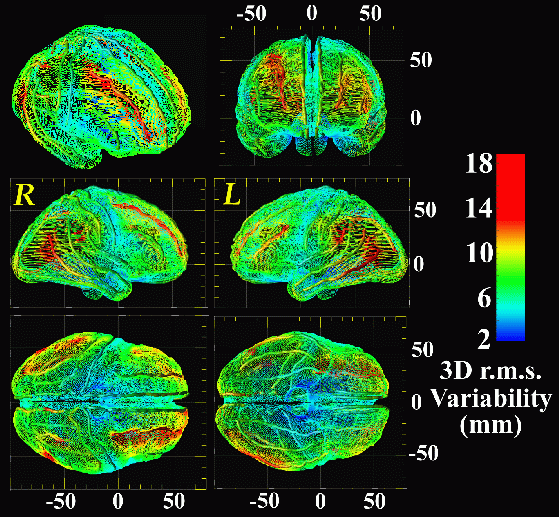
Anatomical Variability of the Cerebral Cortex. (N=9, Alzheimer's Patients). A 3D map of variability is shown (above), on an average surface representation of the cortex derived from an Alzheimer's Disease population. Note how the structural variability of temporo-parietal and frontal association areas (red colors) is a factor of ten higher than sensorimotor cortex (blue colors). Variability is calculated based on 3D displacement maps, which locally encode the amount of deformation required to drive each subject's gyral pattern into exact correspondence with the average cortex for the group (below).
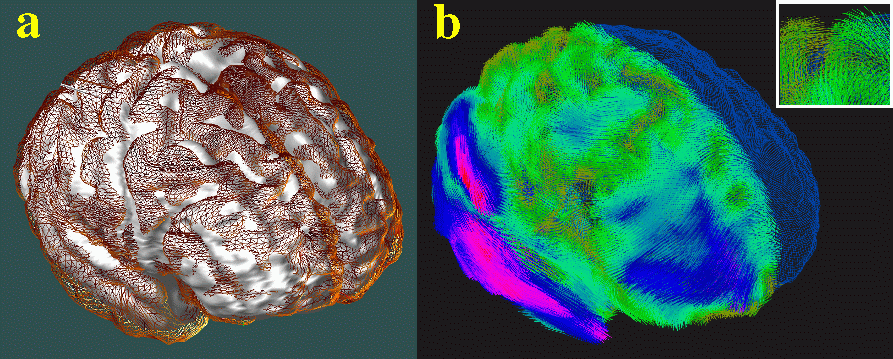
Atlas Construction. High-resolution 3D (256x256x124 resolution) T1-weighted fast SPGR (spoiled GRASS) magnetic resonance imaging (MRI) volumes were acquired from 26 subjects diagnosed with mild to moderate Alzheimer's Disease (NINCDS-ADRDA criteria), and 20 control subjects matched for age, gender, educational level and handedness. Connected systems of parametric surface meshes [1] were used to model 84 structures per brain: 16 deep sulcal, callosal and hippocampal surfaces, all major cortical sulci, Sylvian fissures, 14 ventricular regions, and 36 gyral and cytoarchitectural boundaries in 3 dimensions. 3D patterns of structural variability and asymmetry were analyzed by digitally mapping all structures from all subjects into four different stereotaxic systems, using Talairach [2], affine [3], polynomial [4] and continuum-mechanical mappings [5-8; cf. 9]. When applied to the image data before intensity averaging, these mappings produced a series of imaging templates with increasingly well-resolved anatomical features (below). High-dimensional volumetric maps elastically reconfigured the anatomy of each subject into structural correspondence with a disease-specific average anatomical template. Resulting information on variations in gyral and subcortical topography was encoded as a non-stationary Gaussian random tensor field and visualized in 3 dimensions. Tensor field corrections based on Christoffel fields and Winslow theory were applied to create a crisp average image template with the average anatomical configuration and intensity for the group. Continuum-mechanical matching of cortical patterns enabled the construction of a well-defined 3D average cortex and gyral pattern for AD subjects, and a comprehensive imaging atlas to accelerate future dementia studies.
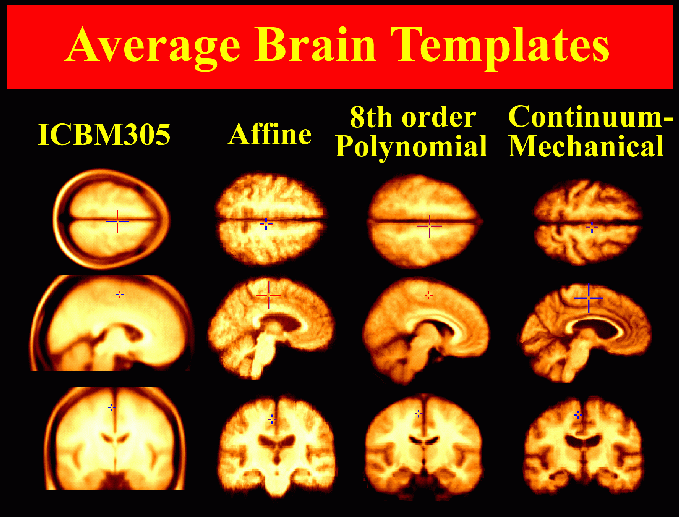
Average Brain Templates. Average brain images can be created by averaging the intensities of MRI scans from multiple subjects. Here, increasingly complex spatial mappings are applied to reconfigure each subject's anatomy into the average anatomical configuration for the Alzheimer's patient population, before the intensities of individual scans are averaged across the group. The final image template (last column), which is based on continuum-mechanical mappings with 0.1 billion degrees of freedom, contains well-resolved anatomical features and represents the average anatomy for the Alzheimer's Disease patient group.
Results. A mosaic of variability patterns emerged, which segregated sharply according to the functional specialization of each system. Sharp contrasts in variability and asymmetry were observed between hippocampal and periventricular anatomy, and between heteromodal and idiotypic cortex.
Fundamental patterns of structural variability in the Alzheimer's disease brain were identified. From an imaging standpoint, registration algorithms were evaluated to determine their ability to reduce anatomic variation across the 84 structures. By tuning algorithm parameters and characteristics of the target atlas template, criteria were defined that allowed anatomic variability to be maximally factored out, allowing functional data to be analyzed in a framework that reflected AD morphology. The resulting probabilistic atlasing strategy is currently being applied to additional dementia, schizophrenia [10], and matched normal populations. This approach may complement other current pattern-theoretic, shape-theoretic and random-field based strategies [q.v., [7]] to detect pathology, encode structural variation, and investigate disease-specific brain structure and function.
References. [1]. Thompson et al., J. Neurosci. 16(13):4261-4274 (1996); [2]. Talairach and Tournoux Brain Atlas (1988); [3-4]. Woods et al., J. Comp. Assist. Tomogr. 16:620-633 (1992) and 22:155-165 (1998); [5-8]: Thompson et al.: [5]. Med. Im. Anal. 1(4):271-294 (1997); [6]. IEEE Trans. Med. Imag. 15(4):1-16 (1996); [7]. Chapter 19, Brain Warping, Toga AW [ed.], Academic Press (1998); [8]. J. Comp. Assist. Tomogr. 21(4):567-581 (1997); [9]. Grenander and Miller, CIS Report on Computational Anatomy (1998); [10]. Narr et al., this volume (1999).
Paul Thompson
| RESUME| E-MAIL ME| PERSONAL HOMEPAGE| PROJECTS |
|---|